Improves the ability to distinguish multiple fluorophores with overlapping emission spectra in biological images of labeled cells by a factor of two or more.
Cells (including normal and tumor cells), tissues, microbial communities, and other biological systems generally consist of multiple interacting components, especially proteins and nucleic acids. In theory, each of these individual components could be labeled with specific probes. In practice, however, the number of identifiable components is severely limited by the difficulty involved in distinguishing different fluorescent reporters with highly overlapping spectra. Linear unmixing with least squares approaches have been applied to spectral imaging to expand the number of distinguishable fluorophores for labeling. While this approach is widely used in commercial instruments and software, the number of distinguishable fluorophores to date has been limited to ~10; while the number of targets in a biological sample may be 100 or more.
This technology is an algorithm that improves the ability to distinguish multiple fluorophores with overlapping emission spectra in biological images of labeled cells by a factor of two or more. To estimate fluorophore concentration, the technology uses two pieces of a priori information about biological fluorescently labeled images. This provides a greater than two-fold improvement in the correct assignment of fluorophores in images of bacteria labeled with known fluorophores, compared to existing methods based on the non-negative least squares approach.
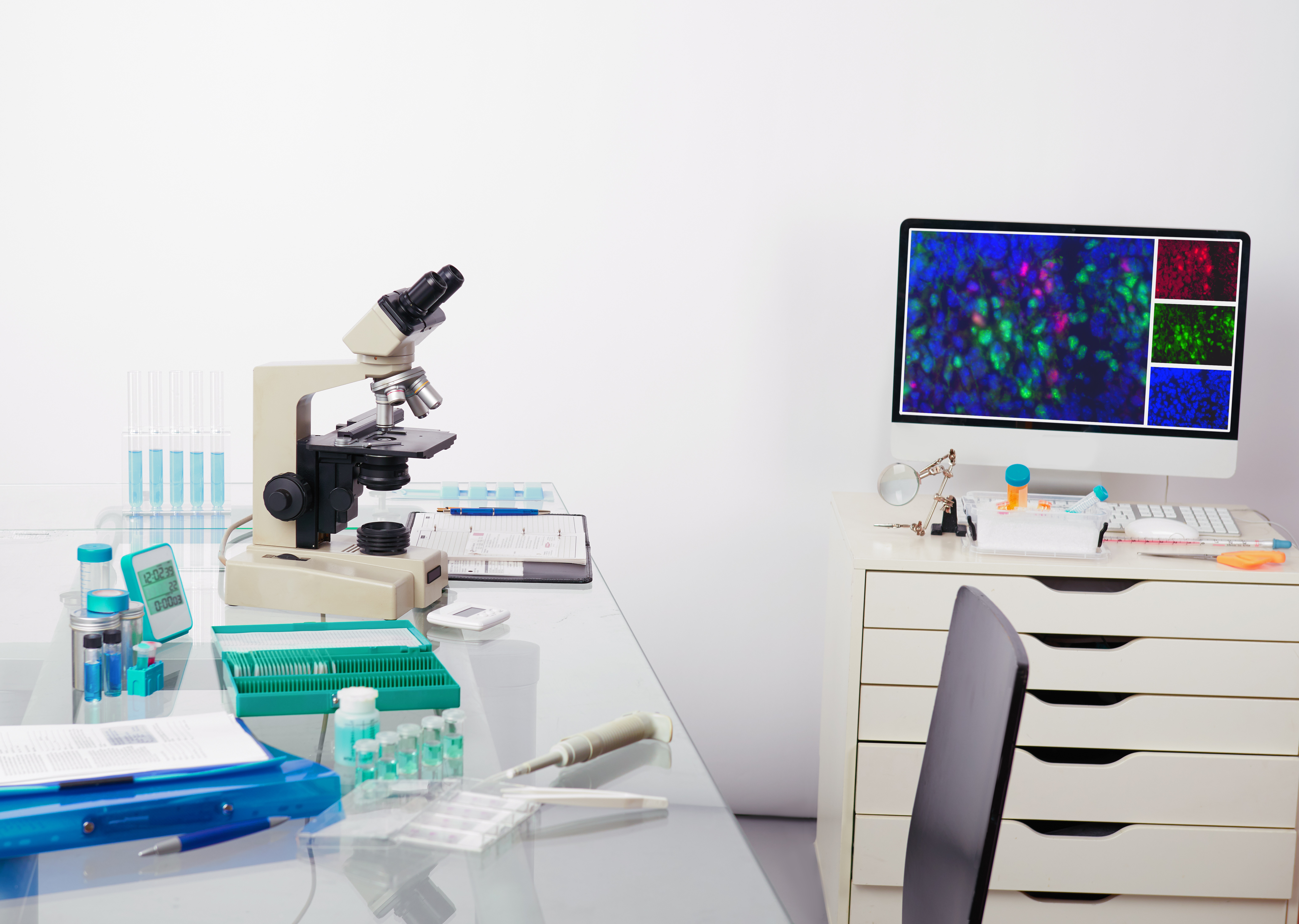
The primary advantage of this technology is its ability to provide a greater than two-fold improvement in the correct assignment of fluorophores in images of bacteria labeled with known fluorophores.
Potential applications for this technology include:
• Biological imaging of complex microbial communities, including the human microbiome.
• Combined labeling in cellular targets: multiple proteins, mRNAs and other biomolecules.
• Clinical research and diagnostic applications including personalized medicine.
Provisional US Patent Application No. 63/441,991 filed 1/20/23.
TRL 3 - Experimental proof of concept
This technology is available for licensing.
This technology would be of interest to anyone involved in research involving the individual components of biological systems, including:
• Educational institutions conducting biomedical research.
• Pharmaceutical developers.
• Medical research laboratories.